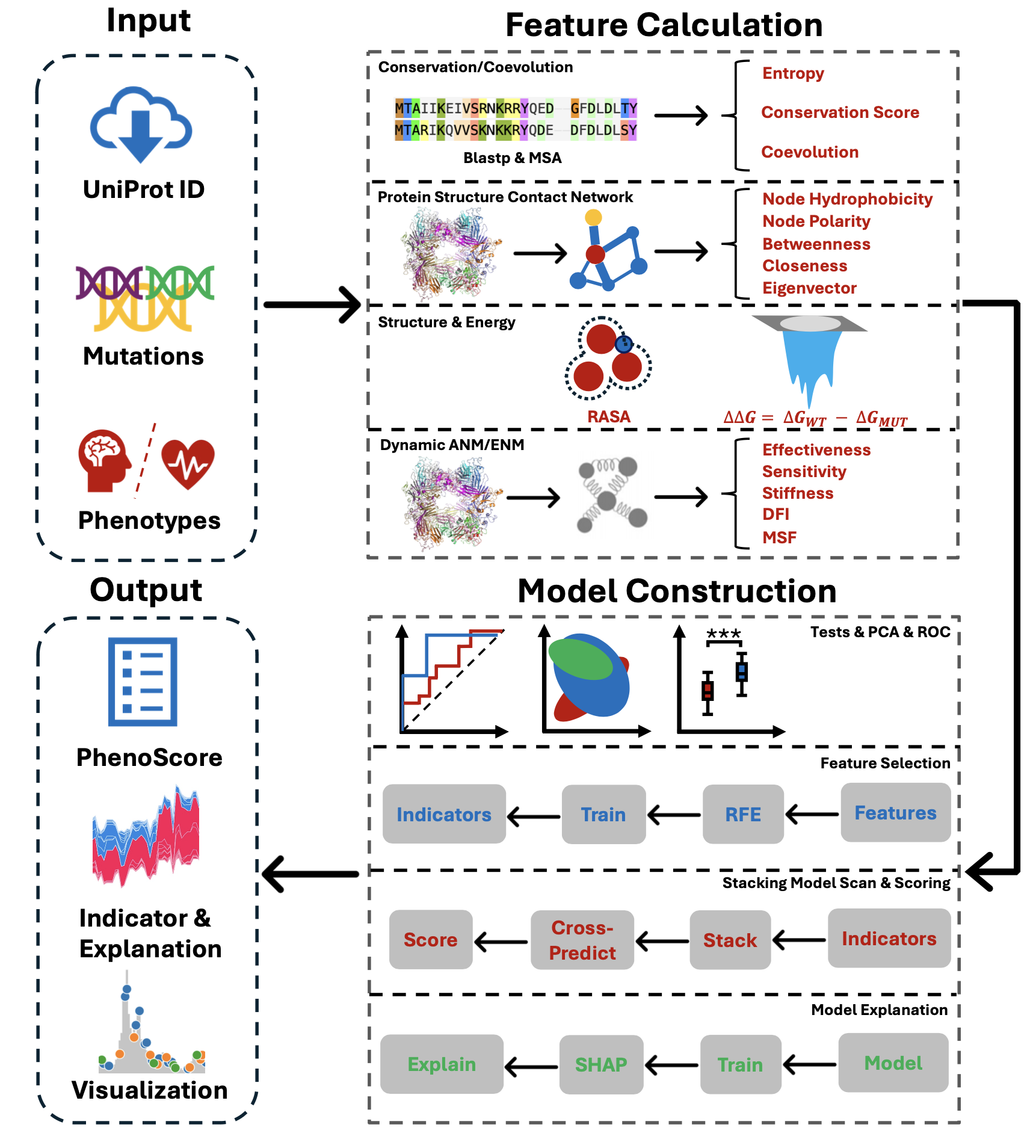
What is protPheMut?
protPheMut (protein phenotypic mutation analyzer) is an online server tool for distinguishing and predicting inseparable single gene missense mutation multiphenotypic diseases. We will automatically calculate the phenotypic mutation characteristics and use the machine learning framework for model training and interpretation. The final results will be presented in a visual result. We will provide biological indicators that distinguish different disease phenotypes caused by single gene missense mutations and mutation score results to measure the biological significance of the current mutation in this phenotype. The network properties of our proteins are calculated based on self-developed NACEN, sequence alignment is based on Blast and Clustal Omega, conservative calculation is based on Rate4site, mutation energy prediction is based on FoldX5 and dynamic network calculation of proteins is based on ProDy. Users need to submit a full-length structure file (or enter the UniProt ID to obtain the AlphaFold prediction results from AlphaFold Database) and records of mutations and corresponding phenotypes, and the returned results include feature results based on different phenotypes, machine learning results, and corresponding mutation scores. Due to the long resource calculation, our Server will prevent other users from submitting files while the Status is running. Please note the Server Status of the Run module. Resources: Download All Dataset